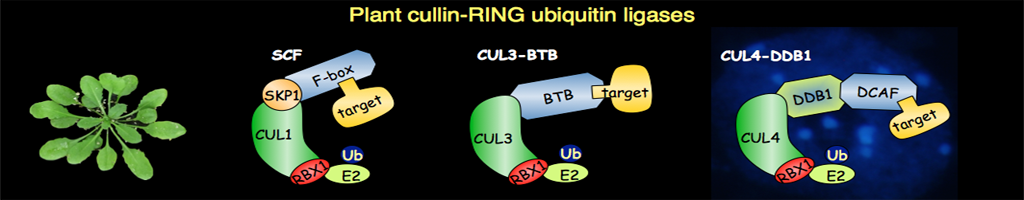
The ubiquitin-proteasome system (UPS) was discovered as a central player for rapid and selective proteolysis in the nucleus and the cytosol. This pathway is also involved in the degradation of short-lived and key regulatory proteins that control a variety of cellular processes, such as the cell cycle and various developmental and hormonal signalling cascades. Degradation via this pathway is a two-step process in which the protein is first tagged by covalent attachment of ubiquitin, and subsequently degraded by the 26S proteasome. Notably some ubiquitylated proteins can also be degraded by selective autophagy. The transfer of ubiquitin to the target proteins requires ubiquitin protein-ligases (E3s), which are the key components in the pathway, as these enzymes determine the specificity of the ubiquitylation reaction.
Recent results have shown that plants make extensive use of regulated proteolysis by the UPS to modulate signal transduction pathways, in particular phytohormones, light signalling and stress responses. On the basis of the Arabidopsis and rice genome sequences, it is predicted more than 1000 E3 ubiquitin protein-ligases. So far, biological functions have only been assigned to a limited number of them. Therefore a better understanding of their function and the identification of their substrates is essential at the fundamental level, but will also be of great value for biotechnology and agriculture.
Our laboratory works on a selected set of Arabidopsis thaliana E3 ubiquitin protein-ligases involved in the cell cycle control, phytohormone signalling and post-transcriptional gene silencing. To unravel their cellular functions, we use a multidisciplinary approach combining, genetics, molecular and cell biology, physiology, biochemistry and structural biology.