RNA degradation stands among the most powerful processes to control gene expression. Diverse and intricate RNA decay pathways cooperate:
- to ensure that the degradation of coding and non-coding RNAs is tightly regulated in response to developmental or environmental stimuli
- to eliminate defective transcripts (RNA quality control)
- to counterbalance loose transcriptional control by degrading transcripts generated from intergenic regions (RNA surveillance)
- to fight pathogens such as viruses.
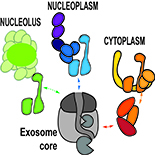
Our main objectives are to identify key actors of RNA degradation pathways in plants, and to determine their impact on genome expression, development or stress response. Our studies currently focus on new co-factors of the RNA exosome, on enzymes that adenylate or uridylate RNAs and on new factors associated to P-bodies and decapping activators. Finally, we address the roles of RNA degradation pathways during viral infections.
Our main experimental strategies include forward and reverse genetics in the model plant Arabidopsis thaliana, protein biochemistry approaches coupled to mass spectrometry analyses, and new high-throughput techniques based on Illumina and Nanopore sequencing to identify 3’ modifications of transcripts.
Key funding of our current research includes the NetRNA LabEx (2011-2028) and the ANR grants 3’modRN (2015-2021) and URIVir (2021-2025).