The platform is specialized in nucleic acids sequencing and genes expression analysis. Strategically, it constitutes a center of expertise in “long reads” sequencing with Oxford Nanopore technology.
The objectives of the platform are to provide public and private research teams with tools and skills to carry out their projects in genomics, transcriptomics and epigenetics. It has new generation instruments for NGS sequencing (Oxford Nanopore and Illumina), high-throughput Crispr-Cas9 genotyping (HRM) and genes expression analysis (qPCR and ddPCR).
The platform is defined according to a charter. It offers services according to a price list, and research collaborations for the implementation of projects with the development of specific methods.
Networks
The platform is a member of Cortecs, a network of scientific research and service platforms of the University of Strasbourg, in partnership with CNRS and INSEM.
The platform labeled IBiSA , a scientific interest group (GIS) which conducts a selective national policy of labeling and support for biology, health and agronomy platforms and biological resource centers (CRB).
Equipments
- Oxford Nanopore Sequencing: PromethION P2 and 2 MinION with Flongle Adapter
- Illumina NGS sequencing: MiSeq
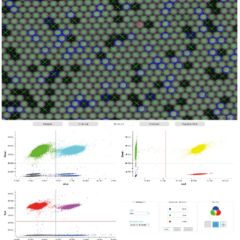
- Digital PCR: Stilla Naica System
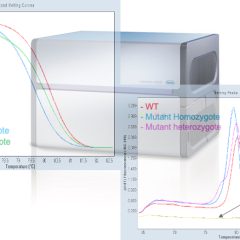
- Genes expression analysis by qPCR and HRM genotyping: Roche LC480-II
- Quality control of NGS sequencing samples and libraries: Bioanalyzer 2100 (Agilent); Qubit (ThermoFisher Scientific)
How to submit a project to the platform?
To submit a project to the platform, contact us by email at ibmp-aeg@unistra.fr
We will respond to you within 7 days to define the best strategy to adopt and the technological approach to consider. An cost estimation will then be drawn up. The deadline for carrying out the experiments will be communicated to you after validation of the samples. In some cases, an exchange with a bioinformatician will be proposed to you before the launch of the project.