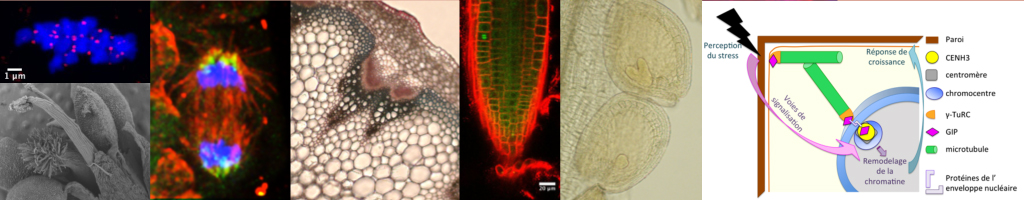
Plants are sessile organisms that are continuously exposed to various stresses (mechanical, abiotic and biotic stresses) during their development. They have evolved mechanisms to regulate their growth and adapt to these stresses, through signaling pathways transmitted to the nucleus.We aim at deciphering these pathways using interdisciplinary approaches (-omics, cellular, physics, genetics and (epi)genomics) through the study of key players located at the nucleo-cytoplasmic interface. We are interested in GIP proteins, regulators of both microtubular networks and nuclear architecture, as well as in regulators of genome integrity maintenance in response to mechanical and osmotic stresses.
Our research programs are partly funded by international (HFSP, IdEX Unistra) and national (ANR) programmes. Our team is part of an European COST network (INDEPTH- Action 16212).