The team research mainly focuses on pentatricopeptide repeat (PPR) proteins, a pivotal class of RNA binding proteins universally found in eukaryotes. Their function is mostly related to the biogenesis of organelles through the fulfilment of essential gene expression processes. Research on PPR proteins has extremely wide implications, going from human health to agriculture. The team contributes to the characterisation of the functional and mechanistic diversity of PPR proteins.
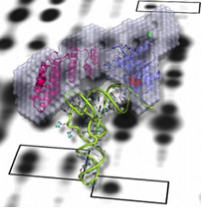
With this research, the team investigates the functional diversity that these essential factors have acquired during the evolution of eukaryotes. In particular, the function of specific PPR proteins called PRORP involved in RNase P activity is explored. Their mode of action is deciphered and interaction networks are determined. Their evolutionary diversity is investigated through the functional characterisation of PRORP in representative model eukaryotes. Because PRORPs have a NYN catalytic domain, we also investigate the diversity of NYN nucleases in plants.
The function of PPR proteins for plant mitochondrial translation is also investigated with a combination of biochemical, structural and reverse genetic approaches. Altogether, this research should help revealing how PPR functions are integrated within the plant cell and how they affect plant physiology.
The team is part of the LabEx consortium MitoCross. It is also funded by different ANR grants.